|
|
|
Restricted Data Portal Added to
GPMDB (2008/11/19)
Since the inception of the GPM system, GPM search servers
have offered investigators the option of storing their results
in the the public data repository GPMDB. Starting today, an
additional choice has been made available for investigators
that would like their results recorded in GPMDB, but would like
to restrict access to those results. This restricted access
option is now available on all search submission pages. Access
to this restricted data can be obtained through the The
GPMDB Portal for Restricted Access Data Sets which can be
used by the investigators for the purposes of manuscript review
or simply to maintain the privacy of their results. A short
description of the portal and its use is available on the GPM
wiki.
|
|
HUPO Plasma Proteome Project 2
Data Release (2008/10/28)
The most recent data from the Human Proteome Organization's
Plasma Proteome Project released into Tranche has been added to
GPMDB, as part of the ProteomExchange system. This release is
comprised of 485 data sets, generated by three laboratories. To
view these data sets, use the data set keyword HUPO
PPP2. To view all data sets identified as being from
plasma, use the Brenda Tissue Ontology accession number for
blood plasma as the data set keyword BTO:0000131.
|
|
New PTM files available
(2008/10/21)
As an ongoing part of the GPM
Tornado project, new post-tranlational modification
annotation files are now available from the GPM ftp
site. These annotation files are based on Release 56.3 of
the SwissProt Protein Knowledgebase and ENSEMBL version 50
protein sequences for the following species:
Homo sapiens
(human_mod.xml);
Mus musculus
(mouse_mod.xml);
Gallus gallus
(chicken_mod.xml);
Rattus norvegicus
(rat_mod.xml); and
Saccharomyces cerevisiae (yeast_mod.xml).
|
|
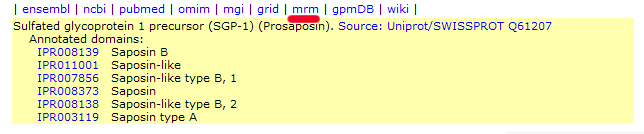
New species included in MRM
worksheet (2008/10/14)
The GPM's Multiple ion Reaction Monitoring Worksheet has
been extended to Mus musculus and Saccharomyces
cerevisiae protein and peptide sequences. The original
release could only be used with H. sapiens sequences.
Links to the appropriate worksheet pages can be found in the
list of links at the top of protein display pages for all
ENSEMBL mouse and SGD yeast accession numbers. These links are
positioned as shown in this example (mouse prosaposin):
|
|
Version 3 of the NCTA protein
lists (2008/10/14)
The 3rd release of the Normal
Clinical Tissue Alliance's tissue specific lists of
observeable proteins is now available. This release includes a
new tissue, liver,
in addition to the original 13 tissue types. The underlying
data has been re-curated and the protein accessions aligned
with the most recent release of ENSEMBL (v. 50).
|
|
Updated versions of distributed
Parallel Tandem for PVM and MPI are now available. (2008/09/08)
Both have been tested for 32 and 64 bit kernels on AMD
Opteron and Power PC (ftp).
This update places together in one distribution the most recent
programs that have been tested for both 32 and 64 bit
compatibility. There are no other new features in this update.
|
|
Changes to the "peptide"
display (2008/09/03)
The display controls for the "peptide" page in the
GPM have been changed to give the user faster access to
information about peptide modifications for a particular data
set. The new control is shown here:
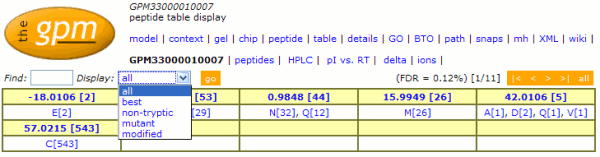
The hyperlinks on the original page that allowed the display
to change from displaying all peptides to either the best,
mutant, non-tryptic or modified peptides have been consolidated
into a single drop-down menu. A new control ("Find")
that allows the display to only show peptides that contain a
particular peptide sequence is located beside this drop-down
menu. A summary table of all of the amino acid residue
modifications that have been assigned in the data set is now
shown just below the new controls. By clicking on the links in
that table, either all of the peptides with a particular
modification mass or the peptides with a particular
mass/residue type combination can be displayed. The numbers in
square brackets are the total numbers of residues modified.
|
|
New X! Hunter spectral libraries
(2008/08/28)
A new set of mass spectrum libraries for use in protein
identifications has been made available. Check the X!
Hunter library page for details. The library curation
process has been improved to improve the accuracy of the
spectra stored in the libraries. Several new species have been
added (horse and fugu), as well as significant expansion of the
zebrafish and arabidopsis libraries because of large new data
sets added to GPMDB.
|
|
X!!Tandem now available
(2008/08/08)
Announcing the availability of X!!Tandem, a version of
X!Tandem parallelized for running on clusters or networks of
workstations.
X!!Tandem is a parallel, high performance version of
X!Tandem that has been parallelized via MPI to run efficiently
on large numbers of CPUs. In X!!Tandem the search is
parallelized by splitting the input spectra into as many
subsets as there are processors, and processing each subset
independently. Both compute-intensive stages of the processing
(initial and refinement) are parallelized, and overall speedups
in excess of 20-fold have been observed on real datasets. With
the exception of minor details related to MPI's program
launching, X!!Tandem is run in exactly the same manner as
X!Tandem, using the same input and configuration files, and
produces exactly the same output.
X!!Tandem is a project of Yale's Keck Biotechnology Resource
Laboratory, and was developed by Robert Bjornson
(robert.bjornson@yale.edu).
The package can be downloaded from here.
|
|
ENSEMBL 50 update (2008/08/01)
The protein sequences in the public GPM have been updated to
the recently released ENSEMBL
50 sequences. These updates also include new SNAP data for
human, mouse and rat sequences.
|
|
Indexing of human, mouse and rat
SNAP data (2008/07/21)
The data collected in GPMDB that demonstrates the presence
of proteins that contain Single Nuclotide-induced Amino acid
Polymorphisms (SNAPs) caused by known non-synomymous SNPs has
been indexed for easier use. In order to find the peptides that
have been observed with an amino acid polymorphism
corresponding to a particular SNP, simply enter the SNP
accession number into the accession
number form (e.g., rs2692696,
rs1800215).
|
|
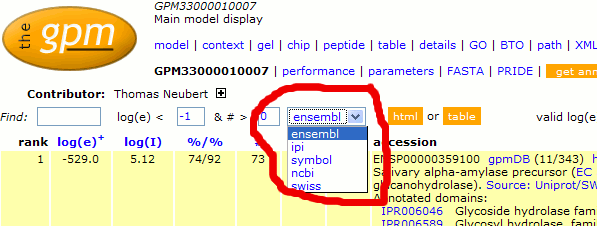
Accession number translation
feature (2008/06/23)
A new feature that allows alternative translations for human
and mouse ENSEMBL protein accession numbers is now available.
Any model originally obtained using ENSP or ENSMUSP accessions
can now be displayed using the following alternative
annotations, without having to search the data again. The
following on-the-fly translations are now available (click on
the link to see the same data annotated with alternative
accessions):
ensembl - default;
ipi - International
Protein Index;
symbol - HGNC
or MGI gene symbols; or
ncbi - NCBI
gene accession numbers;
swiss - Swiss-Prot/Uniprot
accession numbers.
This translation can be performed by selecting the
appropriate value from the "model" page drop-down
list. These translations are approximate, as there may be no
completely accurate translation between various sequence sets,
but the translation is as accurate as we can make it.
|
|
50,000,000th Peptide Id Recorded
(2008/05/16)
Earlier this week, GPMDB passed the 50 million mark for
peptide identifications. We would like to thank all of the data
contributors who have made this project a success.
|
|
Experimental MRM prediction
interface (2008/05/16)
A new interface that attempts to predict multiple-ion
reaction monitoring transitions for peptide sequences has been
made available, for selected human proteins and peptides
(details).
The prediction algorithm is based on the Annotated Spectrum
Library data set, used by X! Hunter for protein
identifications. It attempts to find unique transitions for a
given peptide sequence and parent ion m/z, given parent and
fragment ion mass tolerances. The format of the output page
will change as users give us suggestions to enhance its
utility. The prediction algorithm is available through the new
| mrm
| links that occur on the link toolbars on protein and peptide
display pages.
|
|
Additions to GPMDB interface (2008/05/09)
Several new ways of accessing information in the GPMDB have been added.
Note keywords. A new interface has been added that can query the notes that
data donors have used to annotate individual data sets. The feature can be accessed
using the GPM #'s page, using the
"Note keywords" box.
Multiple peptide sequences. A new interface has been added to the sequence
search page that can be used to determine which peptides of a list have been observed by the GPM. Simply enter the peptide sequences, one
per line, into the box at the bottom of the page.
Human chromosomes. An interface has been created to access all of the human proteins observed
by the GPM organized on a by chromosome is now available here. Clicking
on the desired chromosome will produce the appropriate list. Mitochrondrial and transposon lists are also available through
links below the chromosomes.
|
|
Normal Clinical Tissue Alliance site opens (2008/03/24)
The Normal Clinical Tissue Alliance was formed
to provide information about the proteins that can be observed in clinically derived
tissues. Lists of proteins for each of the tissue types can be obtained and manipulated. The
tissues are "normal" (non-diseased) and the lists represent composites of proteins
obtained from multiple studies. The protein lists are dependent on the availability of
proteomics data: the lists are compiled directly from MS/MS identification data, which is
also made available through the site.
|
|
Change to protein coverage display (2008/03/23)
The protein sequence coverage displays have conventionally showed the regions of a protein that
have been observed in red. The displays have been updated to show the regions of a protein
that are predicted to be difficult to observe (using MS/MS-based proteomics) in green.
|
|
Species added to GPM servers (2008/03/12)
Twenty-two new eukaryote species have been added to the GPM Tornado cloud
server system. These species include 14 new fungi, 2 insects and 6 protists. A list of these new species is
as follows:
- Aspergillus nidulans FGSC A4
- Candida albicans
- Candida glabrata CBS138
- Cryptococcus neoformans var JEC21
- Debaryomyces hansenii CBS767
- Encephalitozoon cuniculi
- Eremothecium gossypii
- Gibberella zeae
- Kluyveromyces lactis NRRL Y-1140
- Magnaporthe grisea 70-15
- Pichia stipitis
- Ustilago maydis
- Yarrowia lipolytica
- Yarrowia lipolytica CLIB122
- Nasonia vitripennis
- Tribolium castaneum
- Cryptosporidium parvum
- Dictyostelium discoideum
- Leishmania infantum
- Plasmodium falciparum
- Theileria parva
- Trypanosoma brucei
These new proteomes were all obtained from NCBI.
|
|
GPM changes to a peer-to-peer grid computing system (2008/02/25)
Starting on Feb. 23, 2008, a peer-to-peer grid computing system (called Tornado) is being rolled out for
the GPM search servers. When a search is submitted to any of the genome, human, mouse or rat search pages, the
system will determine which server on the grid is the fastest and least busy and send your search to that
computer. This system has the effect of increasing the effective capacity of the existing system
by approximately 10-fold.
Note: In order to use this system, the search form must say "GPM Tornado" at the top of the page. If it
does not say Tornado, please press the reload button on your browser to get the most recent version of the
search form.
|
|
KEGG pathways analysis added (2008/01/24)
A new display using the Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic
pathways database is now available for all data sets that have used the ENSEMBL
proteomes for human, mouse, rat or yeast. The new display categorizes the identified
proteins by metabolic pathway, with about 190 pathways available for human proteins.
The pathways are linked through to their KEGG cartoon representations.
|
|
Annotated modifications added (2008/01/20)
Starting with the 2008.02.01 release of X! Tandem, the search engine will now have
the capacity to set the potential modifications being tested on a protein by protein basis.
This new feature can be activated in either the first round of searching or the subsequence
refinement rounds by setting the "Use sequence annotations" control to be "yes".
The way that this new feature works is quite simple. A file is constructed that contains
a list of sequence accession numbers and potential sequence modification specifications, e.g.
several lines from the human modification file look like the following:
<protein label="ENSP00000166244" pmods="79.966331@Y" />
<protein label="ENSP00000350614" pmods="79.966331@S" />
<protein label="ENSP00000372956" pmods="79.966331@S,79.966331@T" />
<protein label="ENSP00000372947" pmods="79.966331@S,79.966331@T" />
<protein label="ENSP00000363773" pmods="15.994915@P" />
The first line indicates that the protein sequence ENSP00000166244 is known to
be tyrosine phosphorylated. If the "Use sequence annotations" feature is turned on,
then that sequence will be tested for tyrosine phosphorylation, in addition to the other
potential modifications you have specified for your search to be applied to all protein sequences.
Similarly ENSP00000350614
will be checked for serine phosphorylation, ENSP00000372956 and ENSP00000372956 will
be checked for phosphorylation at serine and threonine residues, while ENSP00000363773 will be
tested for the possible presence of hydroxyproline residues.
Files of this type have been constructed for human, mouse, rat, chicken and brewer's yeast
proteomes, using publically available annotation sources such as Uniprot and GPMDB.
This capability is now available on all of the public GPM search servers. The annotation files
are available here.
|
|
GPM system updates (2008/01/06)
Over the Holidays, several system changes have been made to the GPM
system and interface pages. While most of these changes are minor, they
do change some default behaviors.
- The "peptide clustering" control on the protein page has been changed to be consistent with the similar control
on the "peptide" page. For both pages, the default setting is now "all", meaning that
all identified spectra will be represented on the page. This change
was made at the suggestion of a number of users, because of some confusion caused by having only
the "best" identification for each peptide sequence shown as the default.
- More information about the distribution of proteins amongst Gene Ontology categories has been added to the "GO"
display, including p-values for enrichment or depletion each category.
- Shading and other small formatting and navigations changes have been made to make the interface as consistent as possible.
|
|
Copyright © 2008, The Global Proteome Machine Organization
|
|

